|
I am a Bioinformatics Scientist at Cedars Sinai Medical Center, focusing on the analysis and integration of multi-omics, single-cell, and spatial datasets to understand disease mechanisms, particularly in Crohn’s disease. My work spans host-microbiome interactions, microbial co-localization, gene regulation, and immune profiling using high-dimensional data, including imaging mass cytometry. I also collaborate on projects addressing key questions in inflammatory bowel diseases to identify novel biomarkers and therapeutic targets. Previously, I worked as Scientist-II, Computational Biology, with the Global Drug Safety Research Evaluation (DSRE) team at Takeda Pharmaceuticals , San Diego. I am collaborating with multidisciplinary teams to expand our predictive and retrospective safety capabilities. My work included leveraging AI/ML approaches for biomarker discovery, developing splicing analysis pipelines for oligonucleotide projects, and enhancing transcriptomic data analysis pipelines to support drug discovery. During my postdoctoral training at Cedars Sinai Medical Center, I worked on SARS-COV-2 Persistence in the GI Tract | Imaging Mass Cytometry Datasets for Bacterial Co-Localization in Crohn's Disease's , Impact of Human Milk Oligosaccharides Gene Expression and Immune Response in Small and Large Intestine, Quantification of Bacterial Growth Rates (PTRs) from metagenomic datasets (*Ongoing). Before joining Cedars, I also worked as a Postdoctoral Associate at the University of Minnesota Twin Cities, where I worked on various projects to understand microbiome-host interactions in animal and human models under diverse physiological conditions. Which includes Primates Gut Bacteriome , Primates Gut Mycobiome , Gut microbial functional adaptations , Oral microbiome in ( oral/head and neck cancer (HNSCC) , and ex-vivo dental caries), and Animal microbiome of ( Beef cattle , and Horse). I did my PhD in Computaional Biology from Metagenomics and Systems Biology Laboratory at IISER Bhopal and funded by the MHRD. Resume / Research / Email / Google Scholar / Twitter / Github / Tutorials (Under Prep): |

|
|
Most of my research has focused on developing and implementing advanced computational approaches including AI/ML models to uncover the microbiome's role in the overall growth, health, and well-being of both animals and humans. I have extensively studied diet-microbiome-host interactions using multi-omics data. Below is a list of my representative papers. |
|
Jennifer D Cohen, Dahea You, Ashok K. Sharma, Takafumi Takai, Hideto Hara, Vicencia T Sales, Li Wang, Tomoya Yukawa, Beibei Cai, Toxicological Sciences, 2024 paper We evaluated an in vitro panel of 11 ion channel targets linked to seizure risk using automated electrophysiology, identifying Cav2.1 as the most predictive target for seizurogenic activity. The study demonstrates the value of expanding target panels to improve early prediction of seizure risks in drug discovery. |
|
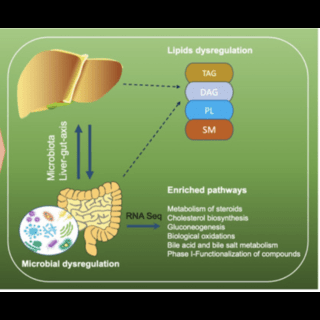
Gaurav V Sarode, Tagreed Mazi, Kari Neier, Shibata, Noreene M, Guillaume Jospin , Nathaniel Harder H.O, AmandaCaceres, Marie Heffern , Ashok K. Sharma, Shyam More, Maneesh Dave, Shannon Schroeder, Li Wang, Janine LaSalle, Svetlana Lutsenko, Valentina Medici Hepatology Communications, 2023 paper Intestine-specific ATP7B deficiency affected both intestinal and systemic response to a high-fat challenge but not the microbiome profile, at least at early stages. |
|
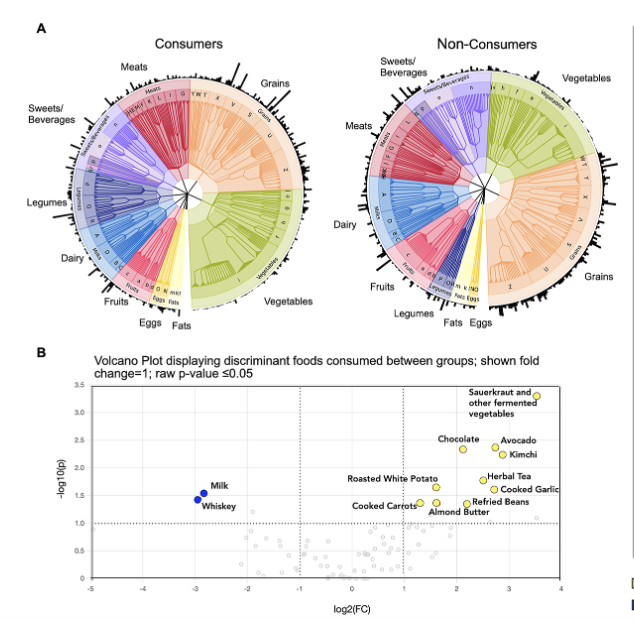
Kylene Guse, Ashok K. Sharma, Emily Weyenberg, Samuel Davison, Yiwei Ma, Abigail Johnson, Chi Chen, Andres Gomez Gut Microbiome, 2023 paper We investigated the impact of lacto-fermented vegetable (LFV) consumption on the gut microbiome and metabolome by comparing faecal microbial diversity, composition, and metabolite profiles between LFV consumers and non-consumers. Results revealed minor microbiome changes but significant alterations in the faecal metabolome, including increased abundances of butyrate, acetate, and valerate, highlighting the need for further research on LFV's health benefits. |
|
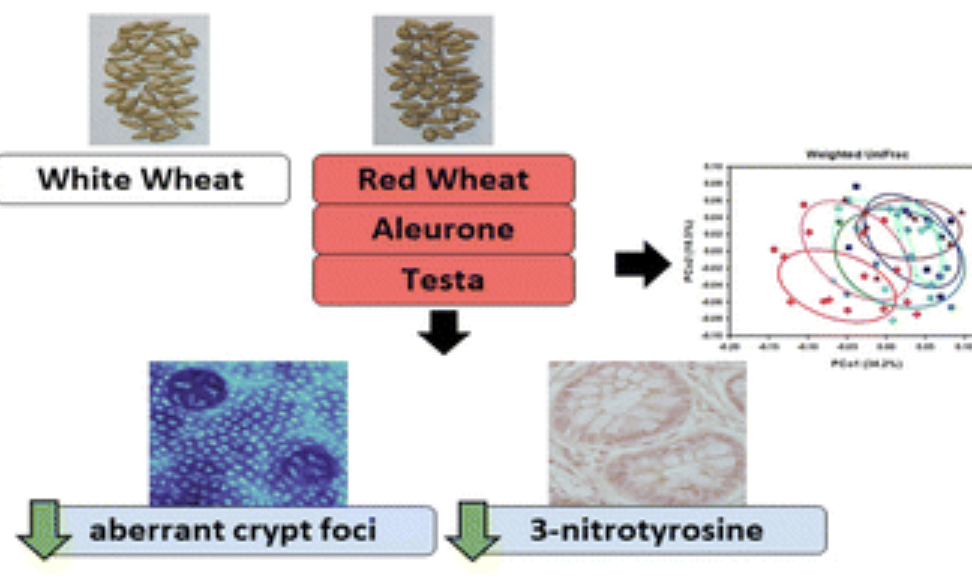
Chelsey Fiecke, Senay Simsek, Ashok K. Sharma, Daniel D Gallaher, Food & Function, 2023 paper We investigated whether the phenolic-rich aleurone and testa layers of red wheat contribute to its chemopreventive effects against colon cancer in a rat model. Red wheat diets reduced biomarkers of cancer risk and altered gut microbiota composition, including increased diversity and changes in key taxa, suggesting the potential roles of dietary fiber and phenolics in reducing colon cancer risk. |
|
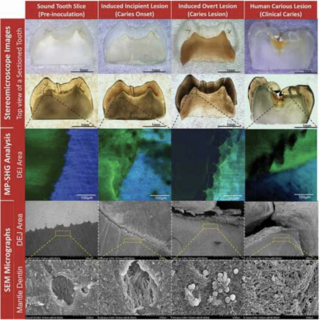
Dina Moussa, Ashok K. Sharma, Tamer Mansour, Bruce Witthuhn, Jorge Perdigao Henriques, Joel Rudney, Conrado Aparicio Badenas, Andres Gomez Journal of Oral Microbiology, 2022 raw data / paper We optimize ex-vivo model integrated with the advanced imaging combined with multi-omics libraries to detect functional signatures associated with dental caries. |
|
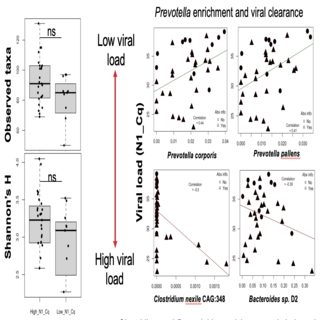
Ashok K. Sharma, Anthony Martin, Jacob Moskowitz, Stephanie Bora, Katherine Lagree, Pieter Dorrestein, David Underhill, Rob Knight, Peter Chen, Suzanne Devkota Gastroenterology, 2022 (Oral Presentation) raw data / presentation / abstract The heavy-dose antibiotic regimen administered to COVID-19 in-patients is associated with viral persistence of SARS-COV-2 in the GI tract, suggesting an important role of the gut microbiome in excluding SARS-COV-2 from the GI tract, perhaps by competitive exclusion or promoting interferon responses. |
|
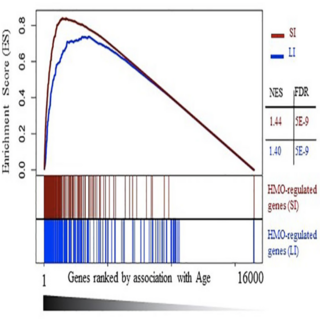
Fernanda Rosa, Ashok K. Sharma, Manoj Gurung, David Casero, Katelin Matazel, Lars Bode, Christy Simecka, Ahmed Elolimy, Patricia Tripp, Christopher E Randolph, Timothy W Hand, D Keith Williams, Tanya LeRoith, Laxmi Yeruva Frontiers in Microbiology , 2022 raw data / paper / project page In this study, we have shown that HMO administration appears to promote immunoregulatory effects at the gene expression level in the absence of gut microbiota. |
|
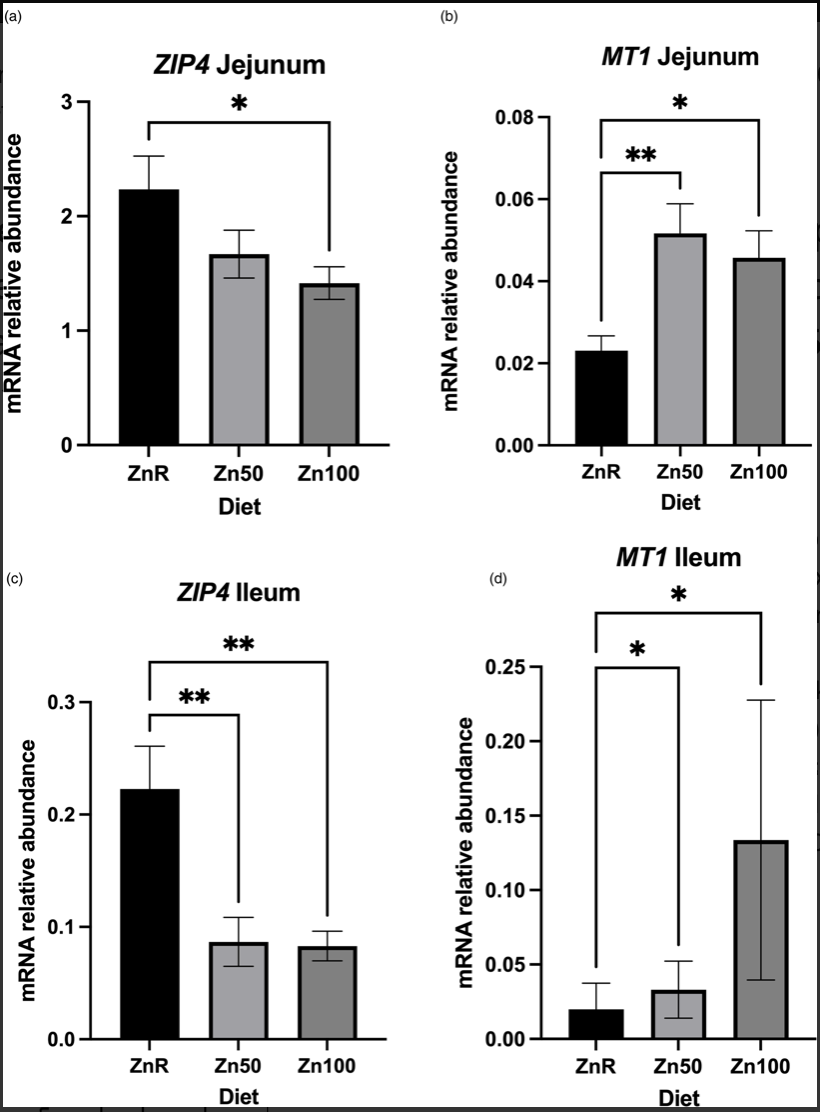
Ramya Lekha Medida, Ashok K. Sharma, Yue Guo, Lee Johnston, Pedro Urriola, Andres Gomez, Milena Saqui Salces Journal of Nutritional Science, 2022 raw data / paper The main goal of the present study was to understand intestinal responses to nutritional Zn restriction and supplementation that could influence overall health of the animal and indicate early changes in response to Zn restriction. |
|
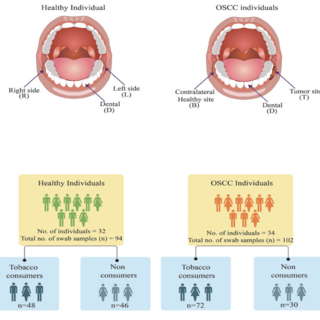
Rituja Saxena, Vishnu Prasoodanan, Sonia Vidushi Gupta, Sudheer Gupta, Prashant Waiker, Atul Samaiya, Ashok K. Sharma, Vineet K. Sharma Frontiers in Cellular and Infection Microbiology , 2022 raw data / paper In this study, we profiled oral microbiome of healthy and oral cancer patients to decipher the microbial dysbiosis due to the consumption of smokeless-tobacco-based products and also revealed the tobacco-associated microbiome. |
|
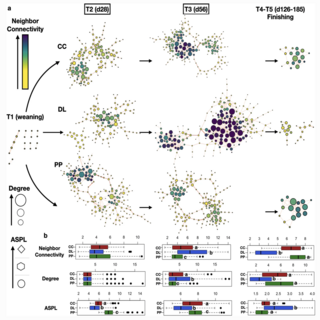
Bobwealth Oakina Omontese, Ashok K. Sharma, Samuel Davison, Emily Jacobson, Alfredo DiCostanzo, Megan Webb, Andres Gomez Animal Microbiome , 2022 raw data / paper This longitudinal study was designed to assess how BKG affects rumen bacterial communities and average daily gain (ADG) in beef cattle. |
|
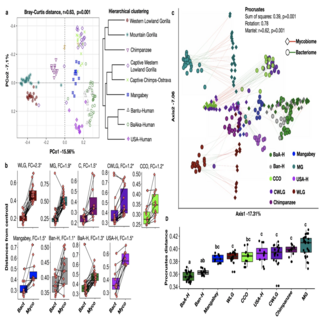
Ashok K. Sharma, Samuel Davison, Barbora Pafco, Jonathan B. Clayton, Jessica Rothman, Matthew McLennan, Marie Cibot, Terence Fuh, Roman Vodicka, Carolyn Jost Robinson, Klara Petrzelkova, Andres Gomez npj Biofilms and Microbiomes , 2022 raw data / paper / project page Our main findings indicate a strong influence of host-ecological factors including diet and lifestyle in shaping the fungal community composition in the primate’s gut. This is in contrast with the gut bacterial fraction, which seems to be more influenced by the host genetics. |
|
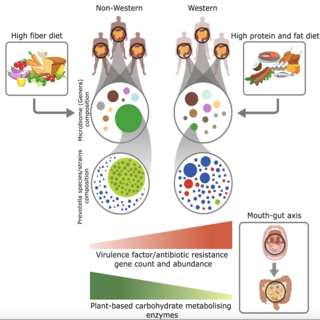
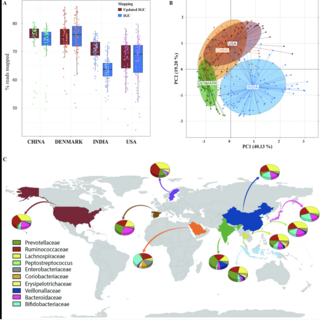
Vishnu Prasoodanan, Ashok K. Sharma, Shruti Mahajan, Darshan Dhakan, Joy Scaria, Vineet K. Sharma npj biofilms and microbiomes , 2021 raw data / paper A population-wide analysis was carried out on 586 healthy samples from western and non-western populations including the largest Indian cohort comprising of 200 samples, and 189 Inflammatory Bowel Disease samples from western populations. |
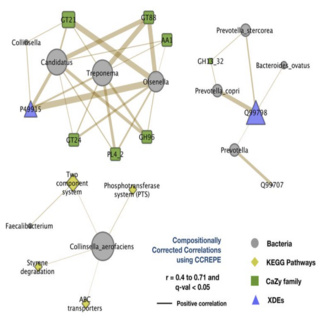
|
Ashok K. Sharma, Klara Petrzelkova, Barbora Pafco, Carolyn Jost Robinson, Terence Fuh, Brenda Anne Wilson, Rebecca M. Stumpf, Manolito Torralba, Ran Blekhman, Bryan White, Karen E. Nelson, Steven Leigh, Andres Gomez mSystems , 2020 raw data / paper / project page The results of this study highlight parallel gut microbiome traits in human and nonhuman primates, depending on subsistence strategy. |
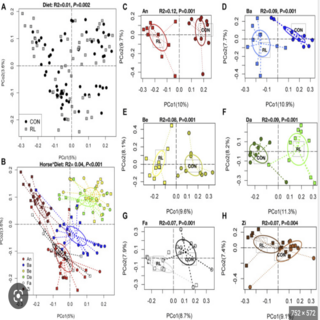
|
Andres Gomez, Ashok K. Sharma, Amanda Grev, Craig C Sheaffer, Krishona Martison Journal of Equine Veterinary Science , 2020 raw data / paper Results in this study indicate that the horse gut microbiome responds in an individualized manner to changes in the amount of acid detergent lignin in alfalfa hay, potentially impacting several feed digestibility characteristics. |
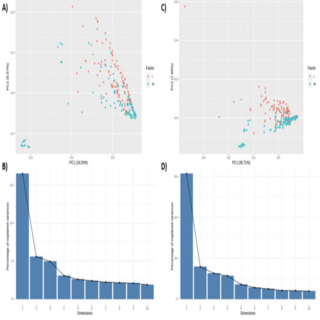
|
Gopal Narayan Srivastava, Aditya Malwe, Ashok K. Sharma, Vibhuti Shastri, Keshav Hibare, Vineet K. Sharma Genomics , 2020 paper / web page We developed the ‘Molib’ tool to predict the biofilm inhibitory activity of small molecules. |
|
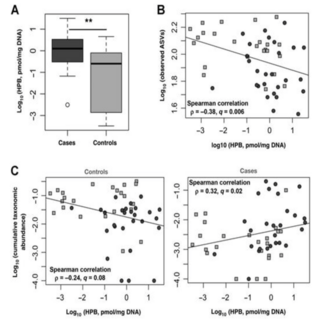
Ashok K. Sharma, William Debusk, Irina Stepanov, Andres Gomez, Samir Khariwala Cancer Prevention Research , 2020 raw data / paper / project page To determine microbial associations with oral/head and neck cancer (HNSCC) among tobacco users, we characterized oral microbiome composition in smokers with and without HNSCC. 16S rRNA MiSeq sequencing was used to examine the oral mucosa microbiome of 27 smokers with (cases) and 24 without HNSCC (controls). |
|
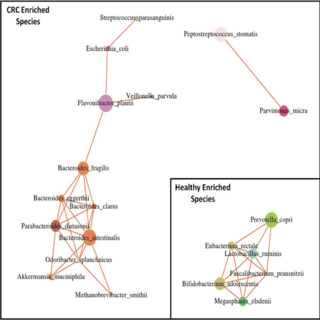
Ankit Gupta, Darshan Dhakan, Abhijit Maji, Rituja Saxena, Vishnu Prasoodanan, Shruti Mahajan, Joby Pulikkan, Kurian Jacob, Andres Gomez, Joy Scaria, Katherine R. Amato, Ashok K. Sharma, Vineet K. Sharma mSystems , 2019 raw data / paper This study provides novel insights on the CRC-associated microbiome of a unique cohort in India, reveals the potential role of a new bacterium in CRC, and identifies cohort-specific biomarkers, which can potentially be used in noninvasive diagnosis of CRC. |
|
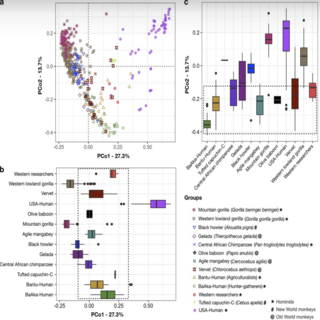
Andres Gomez, Ashok K. Sharma**, Elizabeth K Mallott, Klara Petrzelkova, Carolyn Jost Robinson, Carl J Yeoman, Franck Carbonero, Barbora Pafco, Jessica Rothman, Alexander Ulanov, Klara Vlckova, Katherine R. Amato, Stephanie L. Schnorr, Nathaniel J Dominy, David Modry, Angelique Todd, Manolito Torralba, Karen E. Nelson, Michael Burns, Ran Blekhman, Melissa J. Remis, Rebecca M. Stumpf, Brenda Anne Wilson, Rex Gaskins, Paul A Garber, Bryan White, Steven Leigh mSphere , 2019 paper / project page The results of this study indicate a discordance between gut microbiome composition and evolutionary history in primates, calling into question previous notions about host genetic control of the primate gut microbiome. |
|
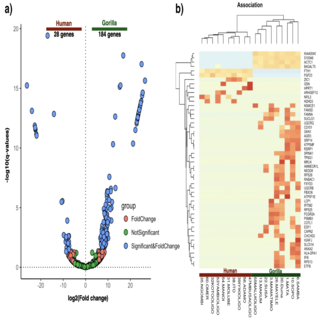
Ashok K. Sharma, Barbora Pafco, Klara Vlckova, Barbora Cervena Jakub Kreisinger, Samuel Davison, Karen Beeri, Terence Fuh, Steven Leigh, Michael Burns, Ran Blekhman, Klara Petrzelkova, Andres Gomez BMC Genomics , 2019 raw data / paper Results in this study suggest that fecal RNA-seq, targeting gastrointestinal epithelial cells can be used to evaluate primate intestinal physiology and gut gene regulation, in samples obtained in challenging conditions in situ. |
|
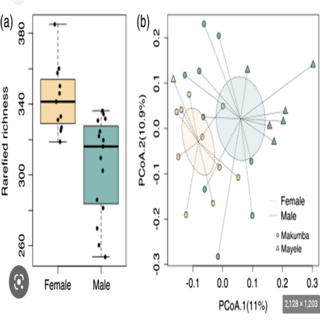
Barbora Pafco, Ashok K. Sharma** Klara Petrzelkova, Klara Vlckova, Angelique Todd, Carl J Yeoman, Brenda Anne Wilson, Rebecca M. Stumpf, Bryan White, Karen E. Nelson, Steven Leigh, Andres Gomez American Journal of Biological Anthropology , 2019 paper Objective of this study was to determine the associations between an individual's age and sex on the diversity and composition of the gut microbiome in wild western lowland gorillas. |
|
Darshan Dhakan, Abhijit Maji, Ashok K. Sharma, Rituja Saxena, Joby Pulikkan, Tony Grace, Andres Gomez, Joy Scaria, Katherine R. Amato, Vineet K. Sharma GigaScience , 2019 raw data / raw data We analyzed the gut microbiome of 110 healthy individuals from two distinct locations (North-Central and Southern) in India using multi-omics approaches, including 16S rRNA gene amplicon sequencing, whole-genome shotgun metagenomic sequencing, and metabolomic profiling of fecal and serum samples. |

|
Subham K. Jaiswal, Ankit Gupta, Rituja Saxena, Vishnu Prasoodanan, Ashok K. Sharma, Parul Mittal, Ankita Roy, , Nagarjun Vijay, Vineet K. Sharma Frontiers in Genetics , 2018 raw data / paper Here, we report the first genome sequence and comparative analysis of peacock with the high quality genomes of chicken, turkey, duck, flycatcher and zebra finch. |
|
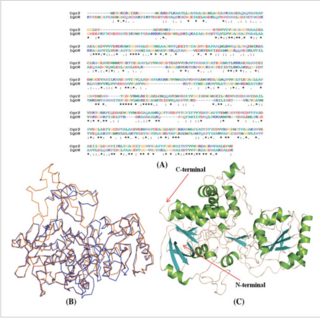
Kundan Kumar, Subham K. Jaiswal, Gaurao Dhoke, Gopal Narayan Srivastava, Ashok K. Sharma, Vineet K. Sharma Journal of Cellular Biochemistry , 2017 paper We report the atomistic details and energy economics binding and metabolism of Digoxin by the Cgr2 protein of Eggerthella lenta DSM 2243. |
|
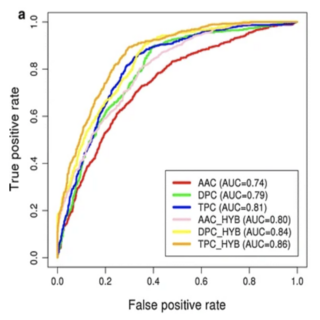
Sudheer Gupta, Ashok K. Sharma**, Vibhuti Shastri, Midhun K. Madhu Dhakan, Vineet K. Sharma Journal of Translational Medicine , 2017 paper / web page In this study, we have developed a prediction tool for the classification of peptides as anti-inflammatory epitopes or non anti-inflammatory epitopes. |
|
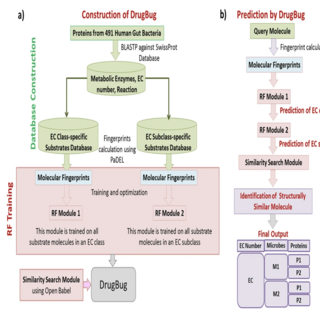
Ashok K. Sharma, Subham K. Jaiswal Nikhil Chaudhary, Vineet K. Sharma Scientific Reports , 2017 paper / web page DrugBug is a computational approach to predict the metabolic enzymes and gut bacterial species, which can potentially carry out the biotransformation of a xenobiotic/drug molecule |
|
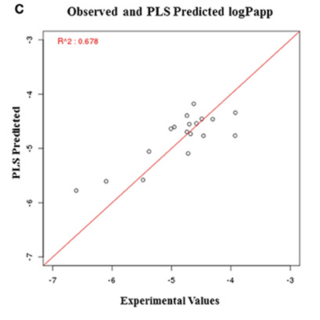
Ashok K. Sharma, Gopal Narayan Srivastava, Ankita Roy, Vineet K. Sharma Frontiers in Pharmacology , 2017 paper / web page ToxiM is a machine-learning based binary classifier for the prediction of toxicity of molecules. Regression models for CaCo-2 permeability and Aqueous solubility has been provided to support the predicted toxicity for a given molecule. For the batch processing please download standalone version of ToxiM from Dataset page. |
|
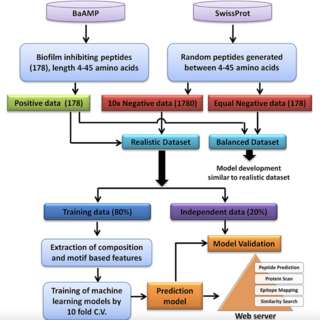
Sudheer Gupta, Ashok K. Sharma, Subham K. Jaiswal, Vineet K. Sharma Frontiers in Microbiology , 2016 paper / web page BioFin is a unique computational method for the prediction of biofilm inhibiting peptides. The experimentally validated biofilm inhibiting peptides sequences and their features were used for the training. |
|
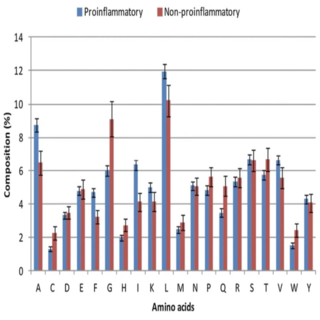
Sudheer Gupta, Midhun K. Madhu Dhakan, Ashok K. Sharma, Vineet K. Sharma Journal of Translational Medicine , 2016 paper / web page ProInFlam is a unique tool for the computational identification of proinflammatory peptide antigen/candidates and provides leads for experimental validations |
|
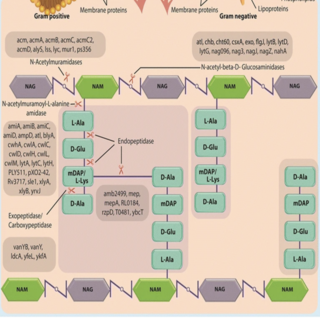
Ashok K. Sharma, Sanjiv Kumar, Harish K., Darshan Dhakan, Vineet K. Sharma BMC Genomics , 2016 paper / web page The present tool helps in the identification and classification of novel peptidoglycan hydrolases from complete genomic or metagenomic ORFs. To our knowledge, this is the only tool available for the prediction of peptidoglycan hydrolases from genomic and metagenomic data. |
|
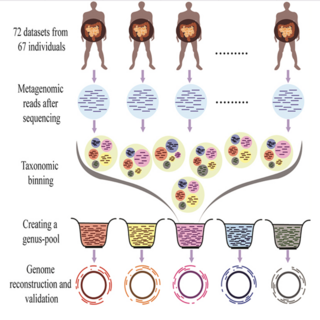
Ankit Gupta, Sanjiv Kumar, Vishnu Prasoodanan, Harish K., Ashok K. Sharma, Vineet K. Sharma Frontiers in Microbiology , 2016 paper In this work, Binning-Assembly approach has been proposed and demonstrated for the reconstruction of bacterial and viral genomes from 72 human gut metagenomic datasets. |
|
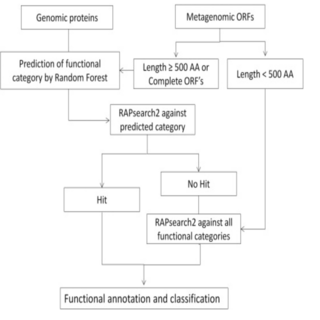
Ashok K. Sharma, Ankit Gupta, Sanjiv Kumar, Darshan Dhakan, Vineet K. Sharma Genomics , 2015 paper / web page Integrated approach for fast and accurate functional annotation of proteins in both genomic and metagenomic datasets. It is available as stand-alone tool as well as a publicly available web-server. |
|
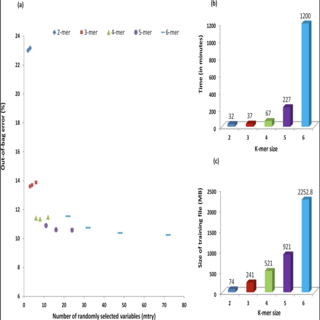
Nikhil Chaudhary, Ashok K. Sharma**, Piyush Agarwal, Ankit Gupta, Vineet K. Sharma PLOS ONE , 2015 paper / web page '16S Classifier' is a Random Forest based tool which is developed to carry out fast, efficient and accurate taxonomic classification of 16S rRNA sequences. It has the unique ability to classify small Hypervariable Regions of 16S rRNA. |
|
Source code credit to Dr. Jon Barron |